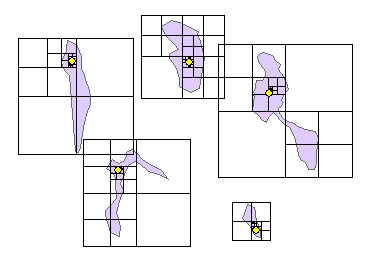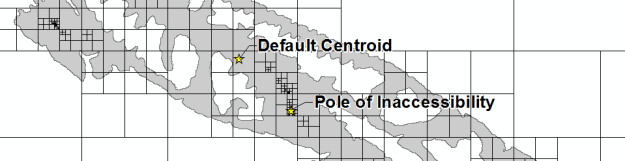
Yesterday, I attempted to replicate the Mapbox Visual Center algorithm using ArcGIS arcpy (Python). Today, I actually looked at the source code (code, license) and attempted to fully port it over from JavaScript. I think it works properly, but you tell me.
def polylabel(polygon, centroid, precision, debug):
cells = []
precision = precision or 1.0
extent = polygon.extent
minX, minY, maxX, maxY = (extent.XMin, extent.YMin, extent.XMax, extent.YMax)
width = extent.width
height = extent.height
cellSize = min([width, height])
h = cellSize / 2
cellQueue = []
x = minX
while x < maxX:
y = minY
while y < maxY: cell = Cell(x+h, y+h, h, polygon) cells.append(cell.geom) cellQueue.append(cell) y += cellSize x += cellSize bestCell = Cell(centroid[0], centroid[1], 0, polygon) numProbes = len(cellQueue) while len(cellQueue): cellQueue.sort(key=lambda cell: cell.d) cell = cellQueue.pop() cells.append(cell.geom) if cell.d > bestCell.d:
bestCell = cell
if cell.max - bestCell.d <= precision:
continue
h = cell.h/2
cellQueue.append(Cell(cell.x-h, cell.y-h, h, polygon))
cellQueue.append(Cell(cell.x+h, cell.y-h, h, polygon))
cellQueue.append(Cell(cell.x-h, cell.y+h, h, polygon))
cellQueue.append(Cell(cell.x+h, cell.y+h, h, polygon))
numProbes += 4
return [bestCell.x, bestCell.y, cells]
class Cell():
def __init__(self, x, y, h, polygon):
self.x = x
self.y = y
self.h = h
self.d = pointToPolygonDist(x, y, polygon)
self.max = self.d + self.h * math.sqrt(2)
self.geom = arcpy.Polygon(arcpy.Array([arcpy.Point(x-h,y-h),arcpy.Point(x+h,y-h),arcpy.Point(x+h,y+h),arcpy.Point(x-h,y+h)]))
def pointToPolygonDist(x, y, polygon):
point_geom = arcpy.PointGeometry(arcpy.Point(x, y),sr)
polygon_outline = polygon.boundary()
min_dist = polygon_outline.queryPointAndDistance(point_geom)
sign_dist = min_dist[2] * ((min_dist[3]-0.5)*2)
return sign_dist
fc = 'BEC_POLY selection'
sr = arcpy.Describe(fc).spatialReference
centroids = []
label_points = []
cells = []
with arcpy.da.SearchCursor(fc,['SHAPE@','SHAPE@XY']) as cursor:
for row in cursor:
best_point = polylabel(row[0],row[1],.75,'#')
centroids.append(arcpy.PointGeometry(row[0].centroid,sr))
label_points.append(arcpy.PointGeometry(arcpy.Point(best_point[0], best_point[1]),sr))
for cell in best_point[2]:
cells.append(cell)
arcpy.CopyFeatures_management(centroids,r'in_memory\centroids')
arcpy.CopyFeatures_management(label_points, r'in_memory\label_points')
arcpy.CopyFeatures_management(cells, r'in_memory\cells')